Cell News 01/2017
13
NIKON YOUNG SCIENTIST AWARD 2017
2 Log (H:L) Rep1
Log 2(H:L) Rep2
-2
-1
0
1
2
3
RatioH/L normalizedRep2
-3
-2.5
-2
-1.5
-1
-0.5
0
0.5
1
1.5
2
2.5
3
RatioH/L normalizedRep1
TRIM56
TRIM65
1
2
-2 -1.5 -1
2
-1
1 1.5
-0.5
-0.5
0.5
0.5
-2
2.5
-2.5
1.5
2.5
-1.5
-2.5
Light (K0)
ΔsopA
Heavy (K8)
SL1344 WT
Cell Lysis
In-solution digest LysC/Trypsin
anti-K(ε)-GG-IP
Strong cation-exchange chromatography
Peptide identification LC-MS/MS
HCT116 infected
with
Salmonella
SopA-dependent ubiquitination sites
a
b
β-helix
domain
N
C
N
C
N-lobe
TRIM56
RING
c
TRIM56
TRIM65
RIG-I
MDA-5
PPP
MAVS
MAVS
MAVS
MAVS
+
+
IKKε
TBK1
P
P
IRF3
P
Type I IFN
SopA
d
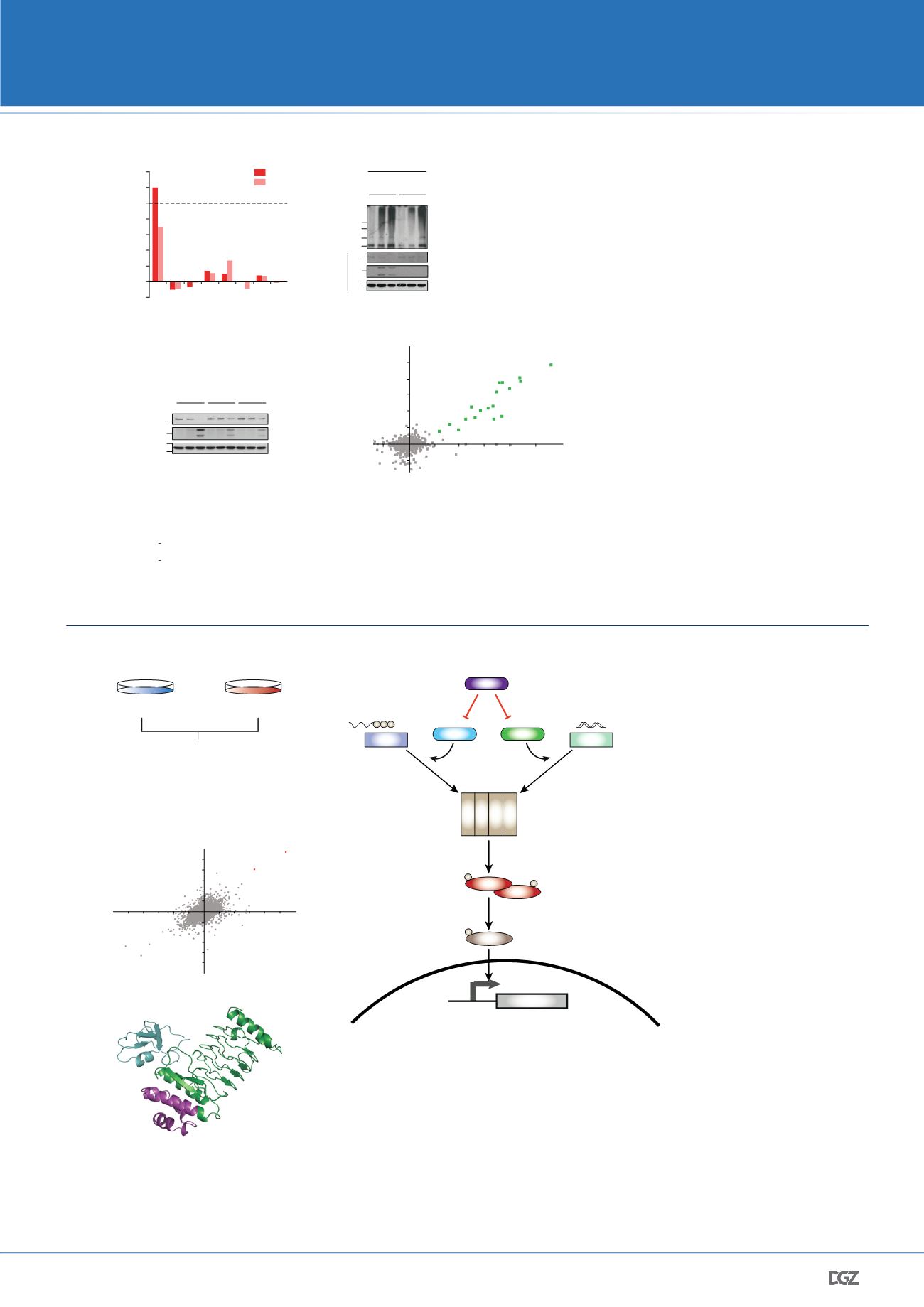
Figure 3: The secreted
Salmonella
E3 ligase effector SopA targets human TRIM56 and TRIM65 ligases to dampen the interferon response upon infection
(a) Design and workflow of the SILAC ubiquitin proteomics experiment used to identify targets of
Salmonella
virulence factor SopA upon infection. (b) Ubiquitin proteomics
recovers human TRIM56 and TRIM65 as targets of SopA. Scatter plot of two replicated experiments displaying log2 (H:L) ratios is shown. (c) Crystal structure of SopA
(163-425) in complex with TRIM56 RING domain. SopA binding is inhibiting TRIM ligase activity (d) Model for inhibition of host interferon signaling by SopA-mediated
targeting and degradation of TRIM56 and TRIM65. Adapted from Fiskin
et al
., 2017
-6
-4
-2
0
2
4
6
RatioH/L normalizedRep2
-6
-5
-4
-3
-2
-1
0
1
2
3
4
5
6
RatioH/LnormalizedRep1
TRAF6
2
4
-4 -3
-2
4
-2
2 3
-1
-1
1
1
5
-5
3
5
-3
BIRC2
TNIP1
IRAK1
TRAF2
TIFA
IKBKG
BCL10
TAB2
RIPK2
RNF31
IRAK4
RBCK1
TANK
TRAF3
N4BP1
ZNRF2
MALT1
R = 0.81
2 Log (H:L) Rep1
Log 2 (H:L) Rep2
-0.2
0.0
0.2
0.4
0.6
0.8
1.0
1.2
1.4
Ex2
Ex1
HCT116 (0.5h)
Log
2
(H:L)
M1 K6 K11 K27 K29 K33 K48 K63
a
(min) 0 15 30 0 15 30
SL1344 WT
∆SopB/
E/E2
IP: M1
Input
IB: M1-Ub
245
135
100
180
IB: IκBα
IB: pJNK
IB: Tubulin
35
48
63
48
b
IB: IκBα
SL1344 (min) 0 15
IB: pJNK
IB: Tubulin
30 0 15 30 0 15 30
Gliotoxin (µM)
0
0.3 0.6
48
63
48
35
d
-4
-5
c
Figure 2: Linear poly-ubiquitination is required for
Salmonella
-induced inflammation
(a)
Salmonella
infection induces the formation of linear M1-linked ubiquitin chains. Log2 (H:L) ratios of all 8 types of ubiquitin chains are displayed. (b) The formation of
linear Ub chains and activation of NF-κB is dependent on the presence of Salmonella virulence factors SopE/E2/B. Lysates from infected epithelial cells were subjected to
immunoblotting using the indicated antibodies. (c) Inhibition of LUBAC activity by Gliotoxin dampens the inflammatory response upon bacterial infection. Lysates from
infected epithelial cells were subjected to immunoblotting using the indicated antibodies. (d) M1-Ub proteomics identifies
bona fide
linear polyubiquitinated proteins.
Scatter plot displaying log2 (H:L) ratios of linear Ub modified proteins recovered upon
Salmonella
infection. Adapted from Fiskin
et al
., 2016


