cell news 2/2013
30
terations of tRNA which is unrelated to the translation of CAG
stretches, tRNAThr-AGU, had no effect (Figure 2B). Interestin-
gly, an increase of tRNAAla-UGC, which pair to the GCA codon
when the ribosome shift the CAG-reading frame to -1 frame,
signifcantly increased the frequency of frameshifting (Figure
2B) suggesting a competition between the glutaminyl-tRNAGln-
CUG and alaninyl-tRNAAla-UGC for the A-site of the translating
ribosome as a mechanism for translational frameshift.
trnagln-cug concentration differs in various cells and
tissues
Clearly, the simultaneous translation of many CAG codons de-
creases the effective concentration of the cognate tRNA leading
to aberrancies in translation. The tRNA sets vary from cell to
cell (6), which raised the question as to whether frameshifting
frequency differs in different cells or tissues. To investigate this
we transfected the Htt51Q(-1)YFP reporter in different HeLa
cell lines that also stably express CFP-tagged variants of Htt,
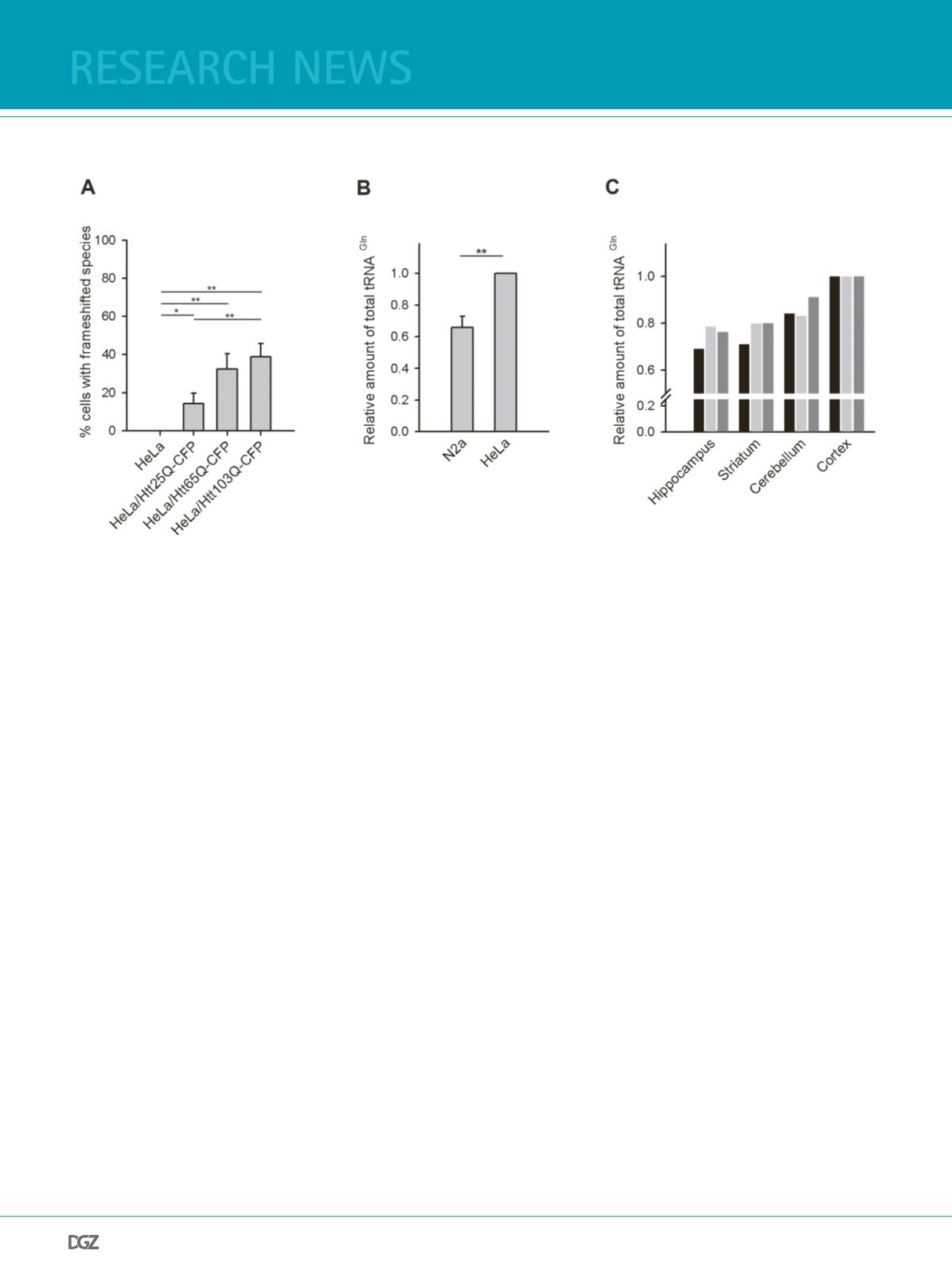
Htt25Q-CFP, Htt65Q-CFP and Htt103Q-CFP (22). We observed
YFP-positive species reporting on the -1 frameshifting (Figure
3A); however the amount of them was much lower compared to
the corresponding N2a cell lines (Figure 1B). Notably, unlike in
the N2a cells, we did not observe any frameshifting in HeLa cells
expression only the Htt51Q(-1)YFP reporter (Figure 3A). Compa-
rison of the tRNAGln-CUG amount in both cells showed much
higher concentration of tRNAGln-CUG in HeLa than in N2a cells
Figure 3. tRNAGln-CUG differs in different cells and tissues:
(a) Quantifcation of frameshifting frequency within the CAG repeat in various HeLa cell lines ectopically expressing Htt51Q(-1)YFP reporter and visualized
after 24 h by fuorescence microscopy (as in Figure 1B). Values are expressed as means of > 5 independent experiments ± SEM. * for p < 0.05 and ** for p<
0.01.
(b) Northern blot analysis of the total tRNAGln-CUG isolated from N2a and HeLa cells. The intensity of the tRNAGln-CUG band is related to the intensity
of 5S rRNA and the values are shown as a mean ± SD of 3 independent experiments. The values of HeLa cells were arbitrarily set as 1. ** for p < 0.01.
(c) Total tRNA concentration in different brain tissues of three mice (differently colored bars for each mice) measured by northern blot. Figure adopted
from (19).
(Figure 3B). Thus the higher frameshifting frequency in N2a cells
cells mirrors the difference in the tRNAGln-CUG concentration
between two cell lines, implying that the tRNAGln-CUG con-
centration is the main cause for frameshifting when translating
extensive amounts of CAG codons. Given the higher vulnerability
of selective loss of the striatal neurons in HD pathology than of
other neuronal tissues, we next asked whether the concentration
of tRNAGln-CUG differs in striatum. Comparison of tRNAGln-
CUG in four neuronal tissues revealed one of the lowest concen-
trations in striatum (Figure 3C). The frequency of the frameshif-
ting in these neuronal tissues is currently under investigation.
translational frameshifting: implications for the hd
pathology
Taken together, our results suggest that simultaneous translation
of large amount of CAG codons leads to aberrancies in transla-
tion (Figure 4). Increased demand for glutaminyl-tRNAGln-CUG
creates a bottleneck which results in -1 frameshifting within the
CAG stretch. Every codon in the CAG repeat is equally susceptible
to frameshifting leading to a formation of a cohort of trans-
frame encoded species with hybrid polyQ/polyA stretches which
differently modulate the conformational switch to nucleate fb-
rillization of the parental polyQ protein (19). This effect strongly
depends on the Q:A ratio generated in each CAG repeat stretch
upon framehsifting (19).
A direct experimental determination of the frameshifting fre-
research news


