cell news 2/2013
31
quency of endogenous Htt is neurons is still missing, but pre-
sence of polyA or polyS proteins in HD-affected brain tissues
(17) and the intrinsic high propensity of frameshifting in neurons
(23) support the notion, that frameshifting may accompany HD
pathology. The striatum is an early target of HD, and striatum has
one of the lowest tRNAGln-CUG concentrations among the brain
regions of mouse. Furthermore, the CAG repeats have the highest
instability in striatum; the CAG stretches are by approximately
10 codons larger than the CAG repeats in any other tissue of the
same individual (24). Thus, is it conceivable to think that the
combination of expanded CAG stretches and the lower tRNAGln-
CUG household would potentiate the frequency of frameshifting
of expanded CAG repeats in striatum. Thus, frameshifting within
expanded CAG stretches may act as modifer of HD pathology
and may contribute to the heterogeneity in disease course and
onset on both cellular level and single individual.
References
1. Plotkin JB & Kudla G (2010) Synonymous but not the same: the causes and consequences of
codon bias. Nat. Rev. Gen. 12, 32-42.
2. Ibba M & Soll D (2004) Aminoacyl-tRNAs: setting the limits of the genetic code. (Translated
from eng) Genes Dev. 18, 731-738.
3. Dittmar KA, Mobley EM, Radek AJ, & Pan T (2004) Exploring the regulation of tRNA distribu-
tion on the genomic scale. J. Mol. Biol. 337, 31-47.
4. Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular orga-
nisms. Mol. Biol. Evol. 2, 13-34.
5. Zhang G, Fedyunin I, Valleriani A, Moura A, & Ignatova Z (2010) Global and local depletion
of ternary complex limits translational elongation. Nucl. Acids Res. 38, 4778-4787.
6. Dittmar KA, Goodenbour JM, & Pan T (2006) Tissue-Specifc Differences in Human Transfer
RNA Expression. PLoS Gen. 2,e221.
Zoya Ignatova did her PhD in Hamburg
with Prof. Dr. Volker Kasche. After her
post-doctoral research at the University
of Massachusetts, she joined the Max-
Planck Institute of Biochemistry in Mar-
tiensried as an Independent Junior group
leader. In 2008 she was appointed as a
Professor in Biochemistry at the Univer-
sity of Potsdam.
research news
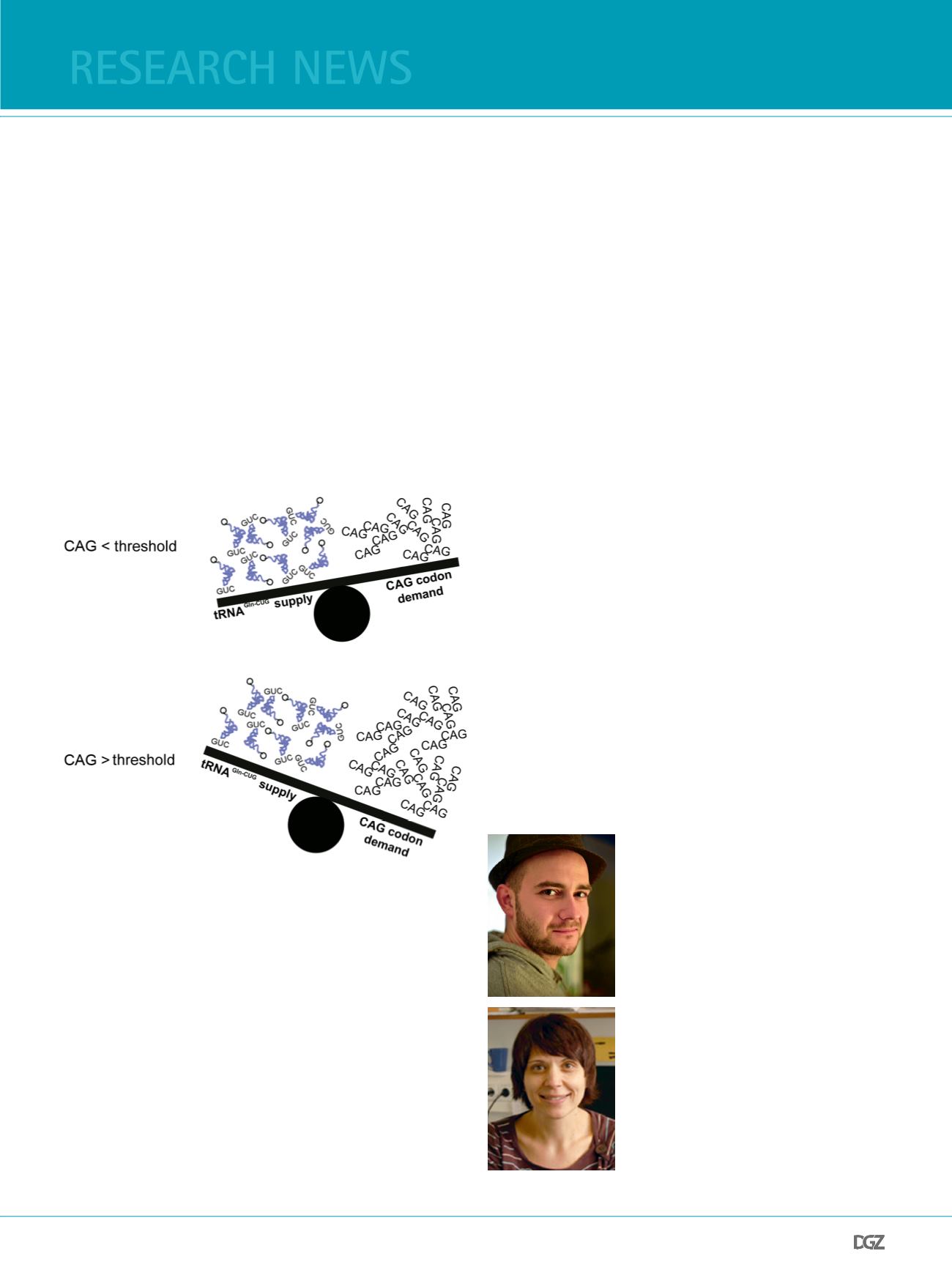
Figure 4. Disbalance between the demand and supply for glutaminyl-
tRNAGln-CUG alters translation of repetitive CAG stretche:
Abnormal expansion of CAG repeats, over the HD disease threshold,
increases disproportionally the demand of glutaminyl-tRNAGln-CUG which
causes translational frameshift within the CAG stretch.
Paul Saffert joined the Ignatova Lab in
2010 for his Master thesis and continued
with the PhD. He studied Biochemistry at
the University of Potsdam where he also
received his Bachelor of Science.
Paul Saffert and Zoya Ignatova:
Biochemistry, University of Potsdam,
Karl-Liebknecht-Str. 24-25, 14476 Potsdam, Germany
Correspondence to: Paul Saffert,
7. Zhang G & Ignatova Z (2009) Generic algorithm to predict the speed of translational
elongation: implications for protein biogenesis. PLoS ONE 4, e5036.
8. Kudla G, Murray AW, Tollervey D, & Plotkin JB (2009) Coding-sequence determinants of
gene expression in Escherichia coli. Science 324, 255-258.
9. Pechmann S & Frydman J (2012) Evolutionary conservation of codon optimality reveals
hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237-243.
10. Zhang G, Hubalewska M, & Ignatova Z (2009) Transient ribosomal attenuation coordi-
nates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274-280.
11. Murakami A, Nakatogawa H, & Ito K (2004) Translation arrest of SecM is essential for
the basal and regulated expression of SecA. Proc. Natl. Acad. Sci. USA 101,12330-12335.
12. Brackley CA, Romano MC, & Thiel M (2011) The dynamics of supply and demand in mRNA
translation. (PLoS Comput. Biol. 7, e1002203.
13. Group THsDCR (1993) A novel gene containing a trinucleotide repeat that is expanded
and unstable on Huntington's disease chromosomes. The Huntington's Disease
Collaborative Research Group. Cell 72, 971-983.
14. Ross CA, et al. (1999) Polyglutamine pathogenesis. Phil. Trans. RS. Biol. Sci. 354, 1005-
1011.
15. Landwehrmeyer GB, et al. (1995) Huntington's disease gene: regional and cellular ex-
pression in brain of normal and affected individuals. Ann. Neurol. 37, 218-230.
16. Orr HT & Zoghbi HY (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575-
621.
17. Davies JE & Rubinsztein DC (2006) Polyalanine and polyserine frameshift products in
Huntington's disease. J. Med. Gen. 43, 893-896.
18. Gesteland RF & Atkins JF (1996) Recoding: dynamic reprogramming of translation. Annu.
Rev. Biochem. 65, 741-768.
19. Girstmair H, et al. (2013) Depletion of Cognate Charged Transfer RNA Causes Transla-
tional Frameshifting within the Expanded CAG Stretch in Huntingtin. Cell Rep. 3, 148-159.
20. Landles C, et al. (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment
that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol.
Chem. 285, 8808-8823.
21. Mangiarini L, et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is suf-
fcient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493-506.
22. Yamamoto A, Cremona ML, & Rothman JE (2006) Autophagy-mediated clearance of
huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 172, 719-731.
23. van Leeuwen FW, et al. (1998) Frameshift mutants of beta amyloid precursor protein and
ubiquitin-B in Alzheimer's and Down patients. Science 279, 242-247.
24. Gonitel R, et al. (2008) DNA instability in postmitotic neurons. Proc. Natl. Acad. Sci. USA
105, 3467-3472.


