Cell News 04/2018
17
BINDER INNOVATION PRIZE 2018
merding 2011). Thus, today’s challenge is not the identification
of force transducing subcellular structures – we essentially
know them already – but elucidating the underlying molecular
processes that regulate force transmission in these complexes.
Understanding the molecular details of mechanotransduction
was complicated by the inability to determine where, when,
and across which molecules mechanical forces are transmit-
ted in cells. We therefore developed Förster resonance energy
transfer (FRET)-based tension sensors that allow the investiga-
tion of molecular force with piconewton (pN) sensitivity in cells
and whole organisms (Grashoff et al. 2010, Krieg et al. 2014,
Austen et al. 2015, Ringer et al. 2017). Here, I will provide a
short introduction into the working principle of this exciting
technology and exemplify how the method can be used to
investigate the inner workings of mechanosensitive complexes.
Working principle of FRET-based tension sensors
The tension sensor technique utilizes Förster resonance energy
transfer (FRET)-based biosensors, in which two fluorophores
undergoing efficient FRET are connected with a mechanosen-
sitive linker peptide (Fig. 1a). This linker element is designed so
that mechanical forces of just a few pN will extend the peptide
and thereby increase the fluorophore separation distance. This
leads to a reduction in FRET, which can be quantified with
fluorescence lifetime imaging microscopy (FLIM). Importantly,
the mechanosensitive linker must fulfil a number of addition-
al criteria to be suitable as a sensor peptide. First, the linker
should not only extend under tension but also refold within a
few milliseconds when forces dissipate (reversibility). Second,
the unfolding and refolding transition pathways of the linker
peptide should be identical (lack of hysteresis) to unambigu-
ously assign a given FRET efficiency to a distinct force value.
Third, the force response should be insensitive to the speed
at which mechanical forces are applied (loading-rate insensi-
tivity), because it is usually unknown how quickly forces act
on molecules in cells. Thus, any tension sensor module (TSM)
needs to be calibrated carefully to ensure that these specific
biophysical requirements are met. The use of non-calibrated
TSMs is strongly discouraged.
We engineered different TSMs, expressed and purified them
from mammalian cells, and used a dual laser trap setup to de-
termine the FRET-force characteristics by single-molecule force
spectroscopy (Fig. 1b). As a result, we established four distinct
TSMs – called F40, FL, HP35 and HP35st – that allow molecular
force measurements at 1–6 pN (Grashoff et al. 2010), 3–5 pN
(Ringer et al. 2017), 6–8 pN and 9–11 pN (Austen et al. 2015).
All sensors are quickly folding, reversible, free of hysteresis and
loading rate insensitive, and therefore suited to study mechan-
ical tension in cells (Fig. 1c). Since our TSMs are genetically
encoded, they can be inserted into proteins of interest to study
force propagation across particular molecules in cells. As the
insertion of the TSM may influence the function of the target
protein, however, extensive control experiments are essential,
as we have extensively discussed before (Austen et al. 2013,
Cost et al. 2015, Freikamp et al. 2016, Freikamp et al. 2016).
Talin sensor measurements provide quantitative
insights into cell adhesion mechanics
Cells’ ability to adhere and sense mechano-chemical signals
is central for numerous biological processes. Much of the
mechanical signals that occur during cell-matrix adhesion are
sensed by and transmitted through a family of cell surface
receptors called integrins (Hynes 2002). Integrins function
as heterodimers, and in mammals 18
α
- and 8
β
-subunits
combine in a restricted manner to form 24 specific receptors
that exhibit specific ligand binding and signaling properties
(Fig. 2a). Each integrin subunit has a large extracellular part
that constitutes the ligand-binding domain, a single trans-
b
a
c
α1 α2
α3
α6
α7
α
v
α5
α8
α4
α9
α10
α11
β2
α
D
α
L
α
M
α
X
αΙΙ
β3 β5
β6
β8
β7
α
E
β4
β1
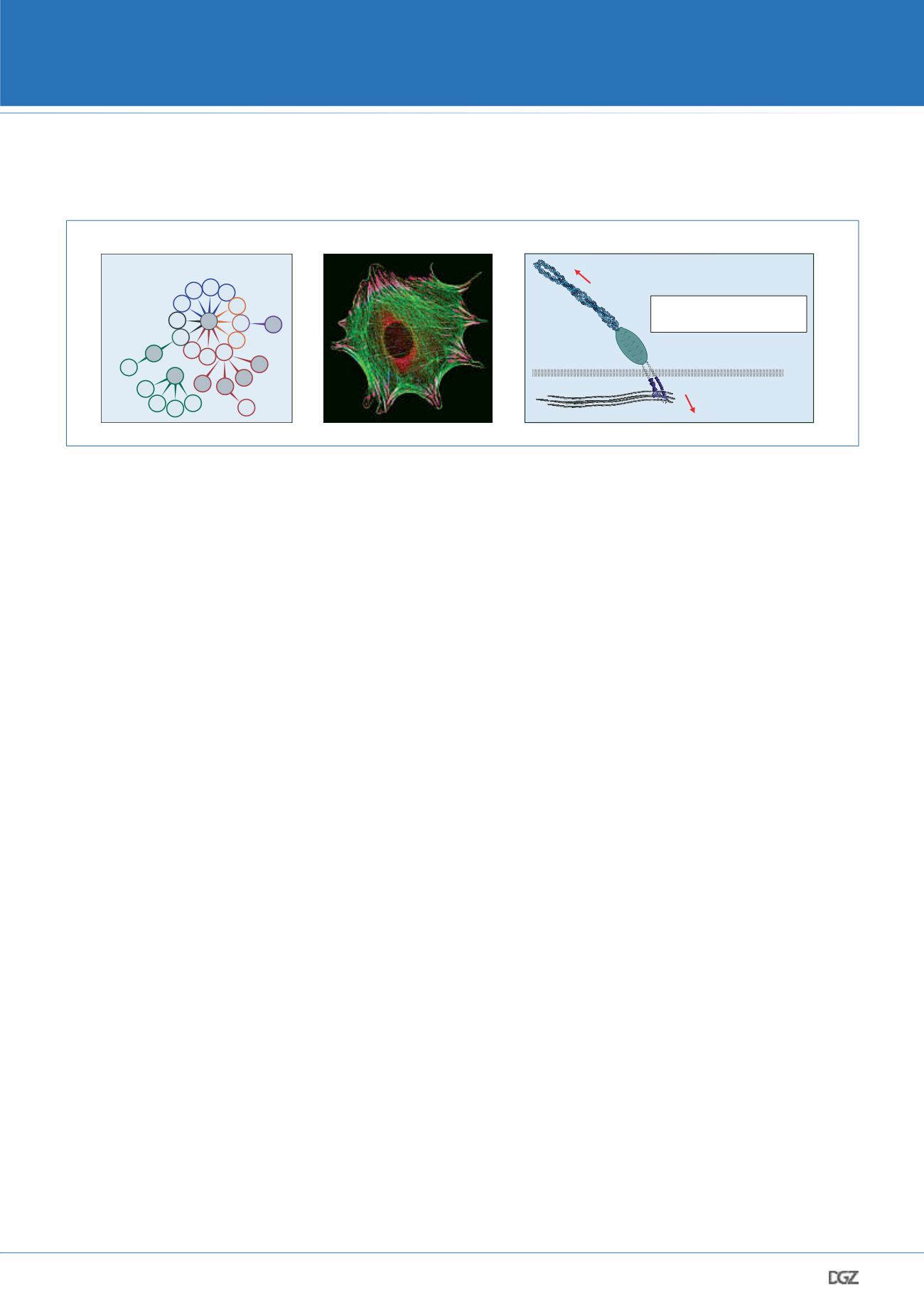
The integrin receptor family
focal adhesion (FA)
f-actin
The cell-matrix interface
ECM
f-actin
>100
FA proteins
integrins
external force
internal force
How do forces propagate
within focal adhesions?
PM
Figure 2: Integrin-mediated cell adhesion. a. Mammals express 24 distinct integrins receptors that differ in their ligand binding and signaling
properties. Red color indicates RGD receptors; blue: collagen receptors; orange: laminin receptors; green: leukocyte-specific receptors. The hemides-
mosomal integrin, which does not require activation through talin, is shown in purple. b. Integrins connect in FAs (pink) to the actin cytoskeleton
(green). c. FAs are macromolecular structures comprising hundreds of proteins. It was unclear how mechanical forces are transduced within these
complexes.


