16
Cell News 2/2015
a near diploid karyotype, but, instead, show a high mutation
rate caused by impaired DNA repair pathways (referred to as
the microsatellite instability or MIN/MSI phenotype). To monitor
microtubule plus end dynamics we expressed the microtubule
end-binding protein 3 (EB3) fused to GFP in mitotic CRC cells
with MIN/MSI or CIN phenotypes and tracked EB3-GFP signals
by live cell microscopy in a time-resolved manner. Intriguingly,
using this live cell assay we found that only cancer cell lines
exhibiting CIN showed significantly increased microtubule plus
end assembly rates during mitosis when compared to MIN/MSI
cell lines, indicating that abnormal microtubule dynamics is in-
deed associated with CIN (Fig. 2A).
Recent evidence indicates that the incorporation of
α/β
-tubulin
subunits into the growing plus end of microtubules is media-
ted by the processive microtubule polymerase
ch-TOG/CKAP5
(Brouhard
et al.
, 2008). Since CIN cells exhibit increased micro-
tubule plus end assembly rates it is reasonable to expect that
lowering
ch-TOG/CKAP5
levels would restore normal microtu-
bule growth rates in those cells. Indeed, expressing shRNAs tar-
geting
ch-TOG/CKAP5
in CIN cells restored normal microtubule
growth rates (Fig. 2B). Moreover, titration experiments using the
microtubule stabilizing drug Taxol showed that sub-nanomolar
concentration of the drug also restored normal microtubule plus
end assembly rates in CIN cells while having only minor effects
on non-CIN cells. Together, these experiments demonstrated
that chromosomally instable cancer cells exhibit an increase in
microtubule plus end assembly, which can be restored by gene-
tic and drug-mediated means.
Increased mitotic microtubule assembly triggers chro-
mosome missegregation and CIN
The observation that CIN cells exhibit an increase in microtubule
assembly and our ability to suppress this phenotype allowed us
to ask whether the abnormal microtubule dynamics can directly
trigger CIN. We used single cell clones derived from MIN/MSI or
CIN cell lines and determined the chromosome number variabi-
lity within a clone after a defined time span of 30 generations
as a measure for CIN. As expected, MIN/MSI cells maintain a
relatively stable karyotype while CIN cells evolve a high karyo-
type variability over time. However, CIN cells, in which normal
microtubule assembly rates have been restored, either by par-
tial repression of
ch-TOG/CKAP5
or upon continuous treatment
with low doses of Taxol, showed a significant suppression of the
karyotype variability (Fig. 2C). These findings clearly establish
a causal relationship between abnormal microtubule dynamics
and mitotic chromosome missegregation and thus, the CIN phe-
notype.
Tumor suppressors and oncogenes that trigger an in-
crease of microtubule plus end assembly during mitosis
It is a big challenge to identify the genetic alterations in cancer
that are responsible for an increase in microtubule growth rates.
However, we established recently a robust assay that allows us
to systematically screen for such regulators in large-scale (Stolz
et al.
, 2015a; Stolz
et al.
, 2015b). As a first step into this exciting
direction we tested the most common genetic alterations found
in human colorectal cancer for their involvement in regulating
microtubule dynamics during mitosis. This candidate directed
approach identified two important lesions that were suffici-
ent to trigger an increase in microtubule assembly rates: first,
the loss of the established tumor suppressor genes
CHK2
and
BRCA1
, which have been previously implicated in DNA damage
response pathways as well as in mitotic spindle assembly (Stolz
et al.
, 2010; Stolz
et al.
, 2011) and second, the overexpression of
the well-known oncogene
AURKA
encoding for the centrosome
associated kinase Aurora-A (Marumoto
et al.
, 2005). Interestin-
gly, we found that the
Chk2-Brca1
tumor suppressor pathway
can act as a negative regulator for Aurora-A kinase activity at
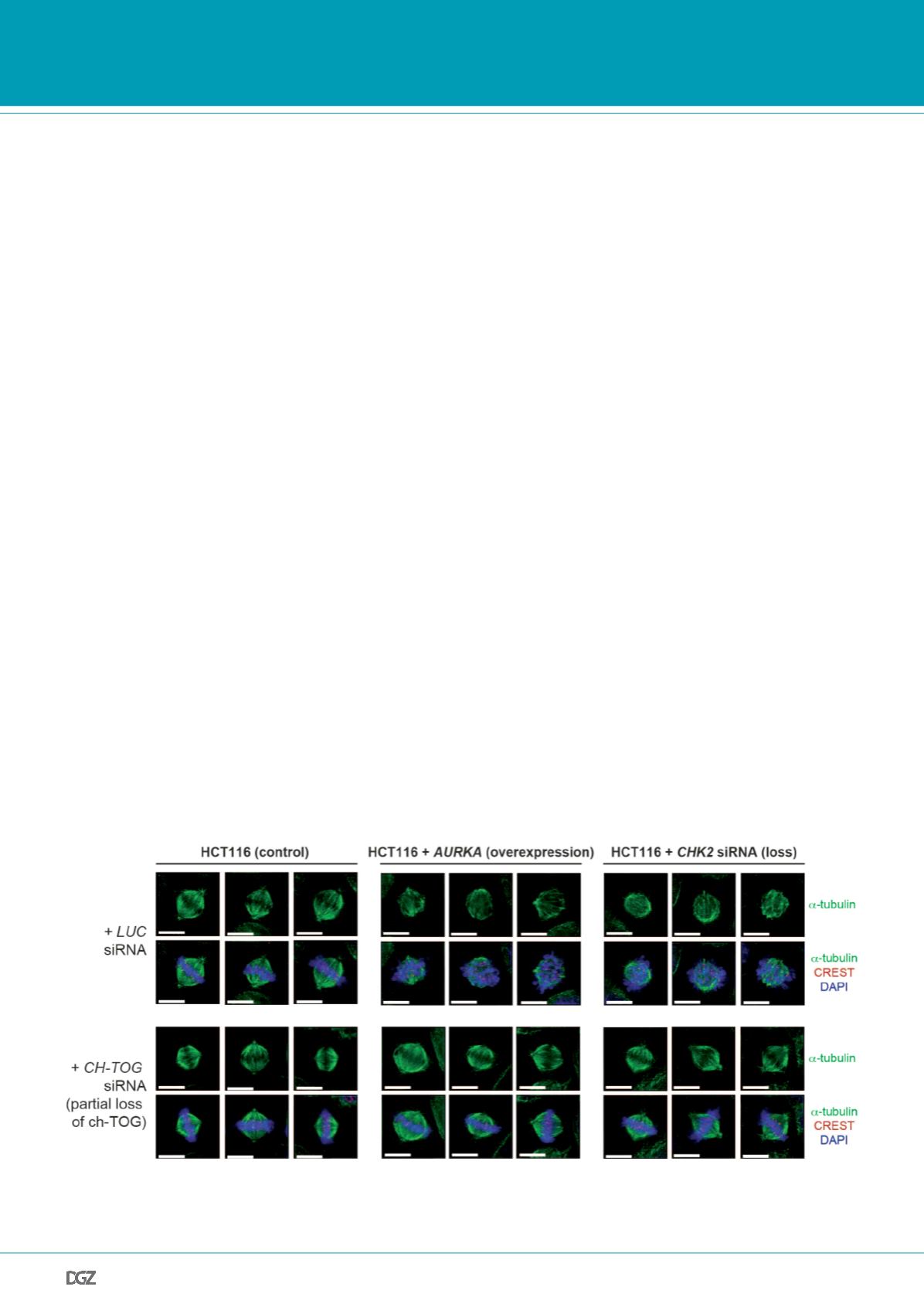
Figure 3:
Increased microtubule assembly rates cause abnormal spindle formation. Human colon cancer cells either overexpressing AURKA or exhibiting
a loss of CHK2, both of which induces an increase in microtubule plus end growth rates, show abnormal spindle geometry and orientation. Examples of
mitotic spindles are given (scale bar, 10 µm). From: Ertych
et al.
, 2014.
PRIZE WINNERS


