cell news 2/2013
16
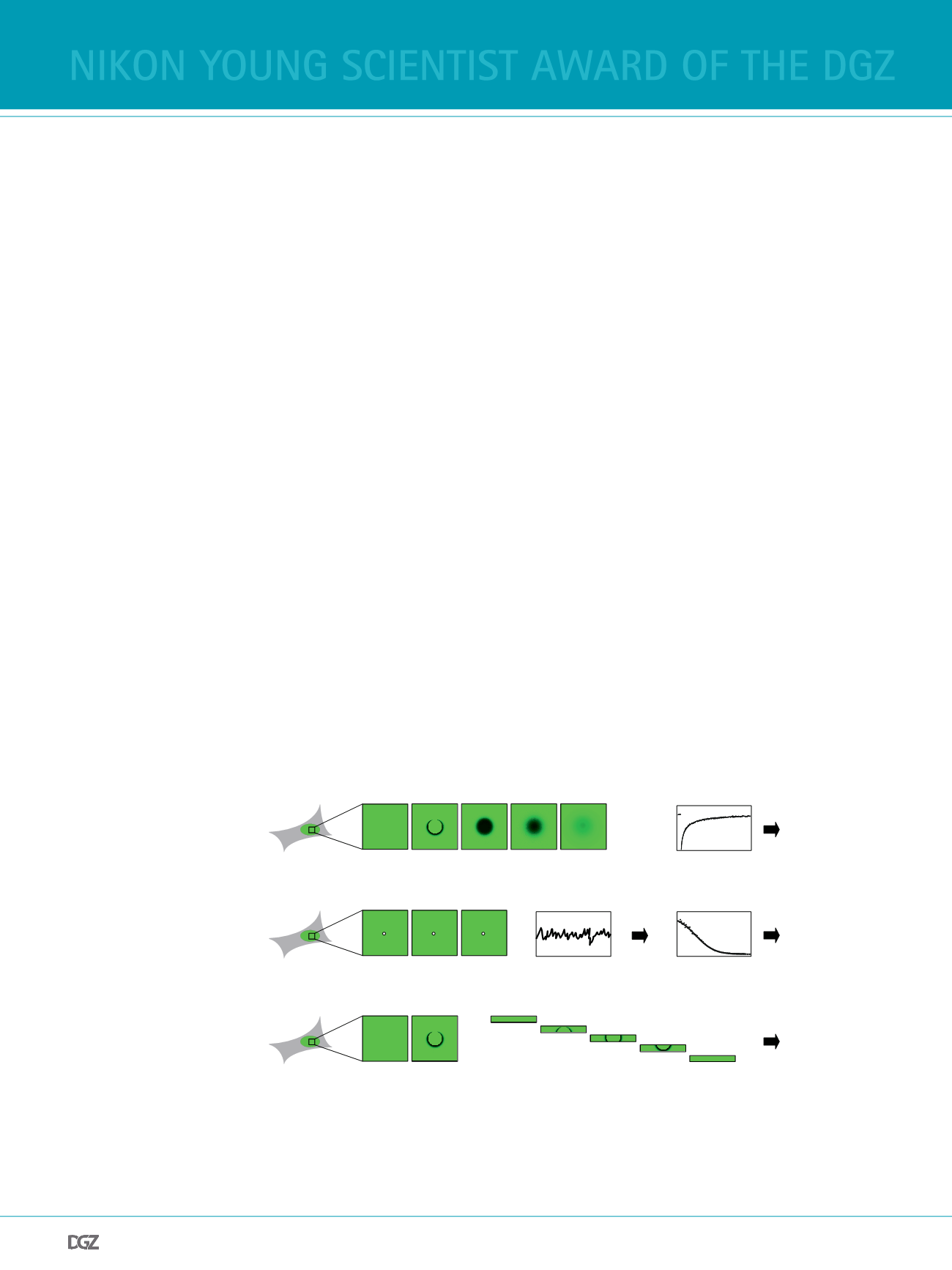
Figure 1 Fluorescence fuctu-
ation microscopy techniques.
Different techniques can be
used to measure the mobility of
fuorescently tagged proteins in
living cells. (A) In Fluorescence
Recovery After Photobleaching
(FRAP), particles within a region
of interest are bleached, and
subsequently the intensity in
the bleach spot is recorded over
time. The shape of the recovery
curve encodes information
about the diffusion coeffcient
of the protein and the kinetic
rate constants for the binding
interactions it undergoes. (B) In
Fluorescence Correlation Spect-
roscopy (FCS), the focal volume
of a confocal microscope is
parked at a fxed position,
and the intensity is measured
over time. When particles enter and leave the observation volume the intensity fuctuates, and the properties of these fuctuations can easily be analyzed
after calculating the autocorrelation function. Fitting of this function yields the diffusion coeffcient and the anomaly parameter that characterizes the
structural complexity of the cellular environment. (C) In Pixel-wise Photobleaching Profle Evolution Analysis (3PEA) particles within a region of interest
are bleached, and the spatiotemporal distribution of bleached particles that move during the bleach process is ftted. This yields information about the
diffusion coeffcient and the rapid binding interactions of the protein under study.
need for speed: tracing chromatin remodelers
in search of the right nucleosome
fabian erdel
In eukaryotic cells, genomic DNA is packaged into a complex
with histone proteins, which is called chromatin. A large por-
tion of the DNA is tightly wrapped around histone octamers to
form nucleosomes, which restrict the access to the underlying
sequence information. Thus, the positioning of nucleosomes is
an important determinant for the accessibility and functiona-
lity of the genome. To actively control nucleosome positions,
cells utilize chromatin remodelers that can translocate or re-
move nucleosomes upon ATP hydrolysis (Erdel et al., 2011a).
Although these enzymes have been studied extensively
in vi-
tro
, their behavior in the context of the physiological chro-
matin template remains poorly understood. In particular, it is
elusive which nucleosome positions are regulated by which
remodeling enzyme, what is the targeting mechanism that
renders a nucleosome a substrate, and how frequently nuc-
leosome translocations occur. To address such questions, it is
instructive to study GFP-tagged remodelers in living cells by
fuorescence microscopy. In particular, fuorescence fuctuati-
on microscopy allows not only to visualize the localization of
the enzymes but provides additional information about their
mobility and the interactions they undergo.
There are several techniques to study the mobility of fuores-
cent proteins in living cells (Erdel et al., 2011b). Most of them
are related to Fluorescence Recovery After Photobleaching
(FRAP) or Fluorescence Correlation Spectroscopy (FCS). In
FRAP, a region of interest is bleached with high laser intensity,
and subsequently an image series is acquired that captures
the motion of the bleached particles out of the bleach region
(Fig. 1A). Based on this image series, the diffusion coeffcient
A
B
C
FRAP (Fluorescence Recovery After Photobleaching)
3PEA (Pixel-wise Photobleaching Profile Evolution Analysis)
FCS (Fluorescence Correlation Spectroscopy)
Fit
Time
Intensity
Time
Intensity
Lag time
Fit
Diffusion coefficient
Binding rates
Diffusion coefficient
Anomaly
Cor
Correlation
Fit
Diffusion coefficient
Binding rates
=
nikon young scientist award of the dgz


